Content
Projects
Goal 1: Roles of Primary Metabolism in Plant Resilience
Plant primary metabolism is closely related with plant growth and development and has great impacts on crop performance including yield, environmental resilience, and nutritional characteristics. Plants need to adjust their metabolism against fluctuating environments to maintain their performance both in short (seconds) and long (generations) time scales. We employ gas chromatography-mass spectrometry (GC-MS) based metabolomics and metabolic flux analysis to elucidate metabolic responses to various environmental stimuli. Our goal is to understand how these metabolic changes contribute to improve plant performance and to identify mechanisms underlying their functions.
Mechanisms of metabolic priming in plant abiotic stress resistance
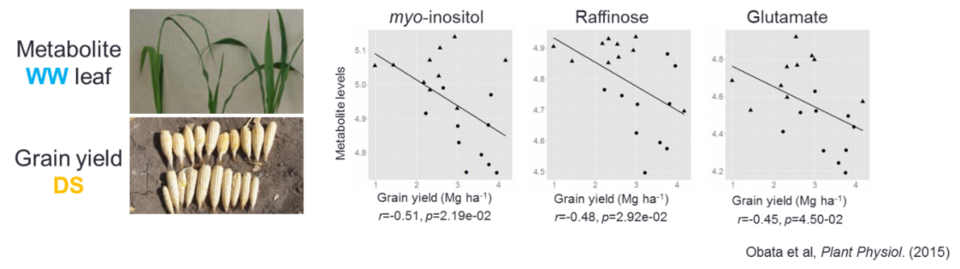
Correlation between leaf metabolite levels under well-watered condition and the grain yield under drought stress condition in 10 tropical maize genotypes (Obata et al. Plant Physiol, 2015)
Plants need to be prepared before the environment changes to unfavorable conditions. There are significant correlations between the metabolic features displayed under non-stressed conditions and plant performance under abiotic stress conditions. One example is from an analysis of metabolite levels and grain yield of maize under drought stress condition in which we revealed the correlation between foliar myo-inositol level in control and grain yield under drought conditions (Fig). This observation together with publications by other authors strongly suggest the importance of basal metabolite profile in plant stress tolerance, which is called “metabolic priming”. Once a metabolite whose level is correlated to stress tolerance is identified, it can be a promising candidate of a biochemical marker for the breeding of stress-tolerant germplasms. Therefore, we are trying to identify metabolites which show correlation with stress tolerance levels by the survey of publicly available data as well as by metabolomics analysis. We are also trying to identify genomic loci related to both metabolic and stress tolerance traits in maize by genome wide association study (GWAS).
Collaboration with Jinliang Yang lab at UNL Agronomy and Horticulture.
This project is funded by Layman Seed Award by UNL (2018-2019)
Identification of the chemical components of soybean seed and sprout related to anti-inflammatory activity
The overall goal of the research team is to generate soybean germplasms for new food products with superior health benefits. To identify the promising target genes to be manipulated, metabolite and protein compositions of the soybean seed and sprout will be characterized and their relationship with dietary anti-inflammatory activity will be examined in the proposed project. Soybean is an excellent source of bioactive peptides and phytochemicals which have exhibited anti-inflammatory activities that play significant roles in preventing obesity and obesity-induced metabolic disorders by dietary intake. Bean sprout is an alternative way of ingestion, which is expected to have further health benefits due to the production of peptides by storage protein digestion and biosynthesis of phytochemicals. In the proposed study, the chemical composition, including phytochemicals and bioactive peptides, of soybean and its sprout will be investigated. These materials will be subjected to the simulated gastrointestinal digestion and, their chemical composition and anti-inflammatory activity on the cultured gastrointestinal epithelial cells will be evaluated. These results will be used to identify the genes regulating chemical components closely related to the health benefits. The proposed research will be the first of its kind to examine the efficacy of germination-derived anti-inflammatory chemicals from soybean to combat and manage the obesity and associated inflammatory responses. The value of food soybean is expected to be improved by proven knowledge regarding the health beneficial effect of soybean consumption and further by generating soybean with greater health benefit.
Collaboration with Kaustav Majumder lab at UNL Food Science
This project is funded by Nebraska Soybean Board (2018-2019).
Goal 2: Establishing the Roles of Multi-enzyme Complexes in Metabolic Network Regulation
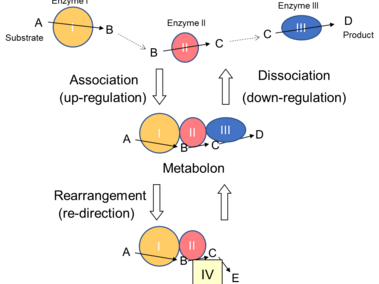

Metabolic regulation by dynamic metabolon (Obata Phytochemistry Rev, 2019).
Rapid, flexible and coordinated regulation of metabolism is essential for all living organisms to keep responding to the environments which fluctuate sometimes within an order of seconds. Currently known molecular mechanisms cannot fully explain this regulatory process. Enzymes catalyzing sequential reactions in a metabolic pathway often form a multi-enzyme complex in which the intermediate metabolites are channeled between enzymes. Theoretical and test-tube experimental studies indicate various advantages of multi-enzyme complex formation in the regulation of metabolic network. However, despite 40 years of research, the functions of multi-enzyme complexes in a living cell are controversial due to unpredictable micro-environment in subcellular compartments. This project will establish functions of a multi-enzyme complex as an additional molecular mechanism regulating metabolism in living cells. This knowledge is crucial to precisely understand how organisms maintain their metabolic homeostasis under fluctuating environments, and what determine organismal metabolic characteristics, which are closely related to organismal performance including productivity and resilience. The knowledge will also be applicable to metabolic engineering. The research results will be disseminated by a newly developed hands-on activity to wide range of audience including undergraduate and high school students, and adult audience. In this activity, participants will play roles of enzymes in a group paper craft activity resembling a metabolic network to intuitively understand the mechanisms of metabolic regulation. This activity will provide a novel, efficient, and interactive education method to improve understanding of metabolic regulation, which will have positive impacts on literacy in metabolism and increase public engagement with metabolic research. Graduate and undergraduate students will be trained their skills in research and public education through both research and education activities.
In this project, the functions of the malate dehydrogenase/ citrate synthase/aconitase multi-enzyme complex of yeast Krebs tricarboxylic acid (TCA) cycle will be investigated as a model system. The research goal of this proposal is to experimentally establish the roles of the TCA cycle multi-enzyme complex in metabolic regulation in living yeast cells. The hypothesis is that the TCA cycle multi-enzyme complex enhances and/or redirects metabolic flux in relation to the ratio of the enzyme complex association. The bi-directional effects between the rate of enzyme association and metabolic flux around the TCA cycle will be assessed through two research objectives. The PI will evaluate 1) the effects of manipulation in multi-enzyme complex affinity on metabolic flux and 2) the effects of metabolic flux changes on the multi-enzyme complex formation. Metabolic flux in the TCA cycle and adjacent pathways will be analyzed by isotope tracer experiments and model-based approaches. Enzyme complex association will be quantified using multiple techniques, including a novel bioluminescence-based realtime assay developed during the project.
This project is funded by NSF CAREER Award (2019-2024; #1845451)